Computation power as you need with EMR auto-terminating clusters: example for a random forest evaluation in Python with 100 instances
One of the main advantage of the cloud is the possibility to rent a temporary computation power, for a short period of time.
With auto-terminating EMR cluster, it is also possible to use a cluster periodically, for example every month, for a specific big data task, such as updating prediction models from the production.
The cost of using a cluster of 100 quadri-processor instances with 15G RAM (m3.xlarge at $0.266 per hour per instance) for 1 hour will be $26.6.
Let’s see in practice with the computation of a Random Forest regressor on production data.
Computing a Random Forest regressor (RFR) on a high volume of data on a single computer will require a few days, which is not acceptable for R&D as well as for production : a failure in the process would postpone the update by 5 days. Let’s see how to launch a cluster every month.
In the following case, preprocessed production data will be a 5.6 G CSV file, where each column is separated by a ; character, the first column corresponding to the label to predict, and the following columns the data to use to make a prediction.
To parallelize on 100 (one hundred) AWS EC2 instances, AWS first requires to raise the initial account’s EC2 limit by applying with a form.
Computing a RFR on a cluster with Spark is as simple as with other libraries.
Create a python compute_rf.py file :
# import the libaries
from pyspark import SparkConf, SparkContext
from pyspark.mllib.tree import RandomForest
from pyspark.mllib.util import MLUtils
from pyspark.mllib.linalg import Vectors
from pyspark.mllib.linalg import SparseVector
from pyspark.mllib.regression import LabeledPoint
import sys
# initiliaze the Spark context
sc = SparkContext()
# get the script's argument (the file path to the data)
filename = sys.argv[1]
print "Opening " + filename
file = sc.textFile(filename)
# get header line, and column indexes
headers = file.take(1)[0].split(";")
index_label = headers.index("label")
index_expl_start = headers.index('expl1')
index_expl_stop = len(headers)
# transform line to LabeledPoint
def transform_line_to_labeledpoint(line):
values = line.split(";")
if values[index_label] == "":
label = 0.0
else:
label = float(values[index_label])
vector = []
for i in range(index_expl_start, index_expl_stop):
if values[i] == "":
vector.append(0.0)
else:
vector.append(float(values[i]))
return LabeledPoint(label,vector)
# filter first line, and transform input to a LabeledPoint RDD
data = file.filter(lambda w: not w.startswith(";xm;ym;anpol;numcnt;")).map(transform_line_to_labeledpoint)
(trainingData, testData) = data.randomSplit([0.7, 0.3])
trainingData.cache()
testData.cache()
model = RandomForest.trainRegressor(trainingData, categoricalFeaturesInfo={},numTrees=2500, featureSubsetStrategy="sqrt",impurity='variance')
# does not work : ,minInstancesPerNode=1000)
predictions = model.predict(testData.map(lambda x: x.features))
labelsAndPredictions = testData.map(lambda lp: lp.label).zip(predictions)
testMSE = labelsAndPredictions.map(lambda (v, p): (v - p) * (v - p)).sum() / float(testData.count())
print('Test Mean Squared Error = ' + str(testMSE))Before going further, test that the script works well locally on a small sample of data :
spark-submit --master local[4] compute_rf.py sample.csvA a third step, upload to AWS S3 the CSV data file and the python script file compute_rf.py, so that the cluster will be able to access them.
Last step, create a cluster with an additional step for the computation to execute after initialization :
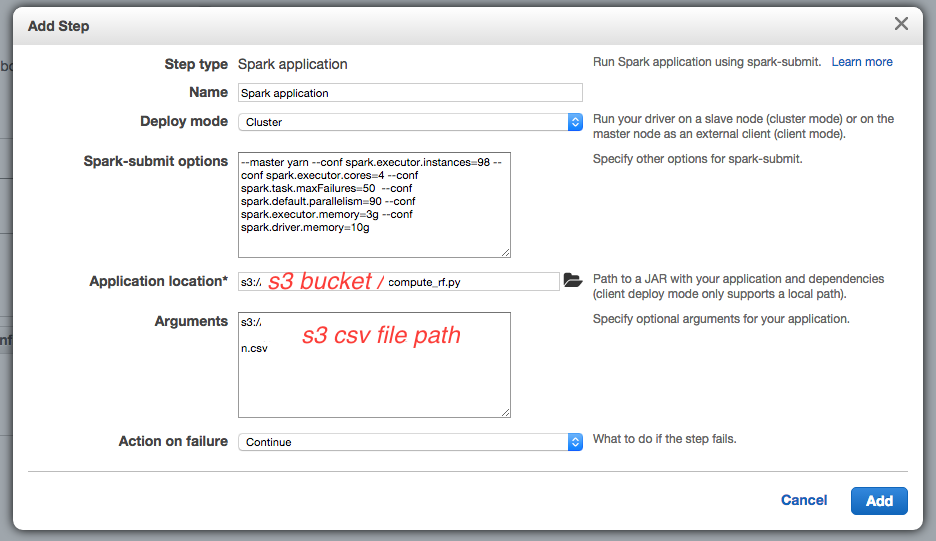
The spark-submit options are :
-
--conf spark.executor.instances=98because I will launch 98 EC2 instances -
--conf spark.driver.memory=10gsince the m3.xlarge instance has 15G memory, and default spark.driver.cores is set to 1, it is possible to give 10G to the driver process on the master instance -
--conf spark.executor.cores=4 --conf spark.executor.memory=3gto use all the 4 cores and 12 G of the 15G of memory of m3.xlarge instances for the slave instances (executors) -
--conf spark.task.maxFailures=50defines the number of failures for a task that will cause the computation to fail, default is 4 which is a bit low. -
--conf spark.default.parallelism=90defines the default minimum number of partitions that will be used in thetextFilemethod : the Spark’stextFilemethod will divide the S3 file into splits that will be read by each executor concurrently. The parallelism should be set to about the number of cores, hence4 x 98 = 392(the program won’t be able to decrease the number of partitions specified here to a lower value) but it might also divide the data file into too small splits, and take more time : one should experience different values for this parameter.
The application location is the S3 path to the Python script. Note that in this version of Spark, you do not need to specify --class org.apache.spark.examples.SparkPi option since Spark will detect the Python file.
The argument is the S3 path to the CSV data file, used in the Python script.
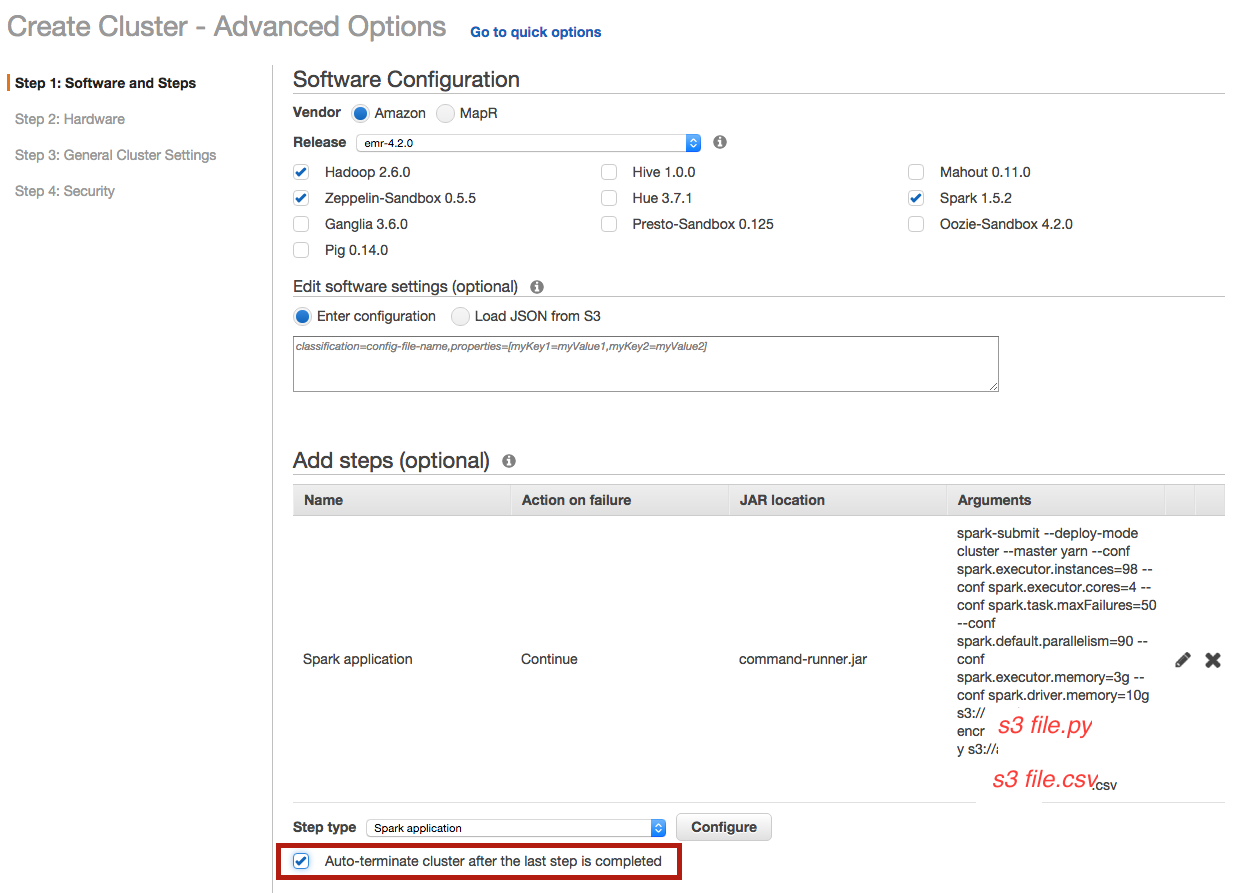
Be careful to select auto-terminating option to close the cluster automatically once the computation is done :
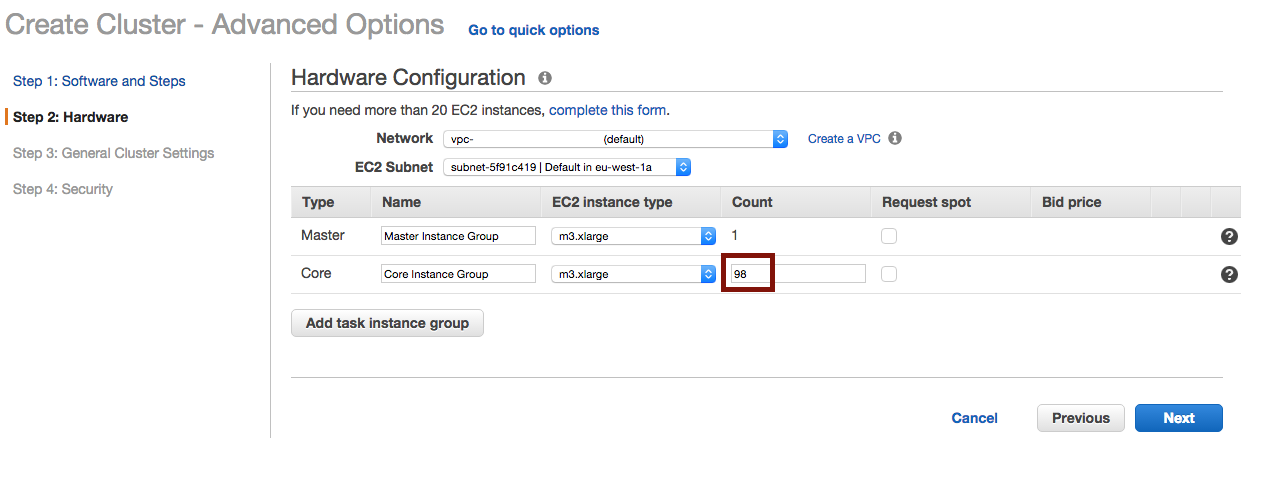
Choose the number of instances :
And define where to output the logs :
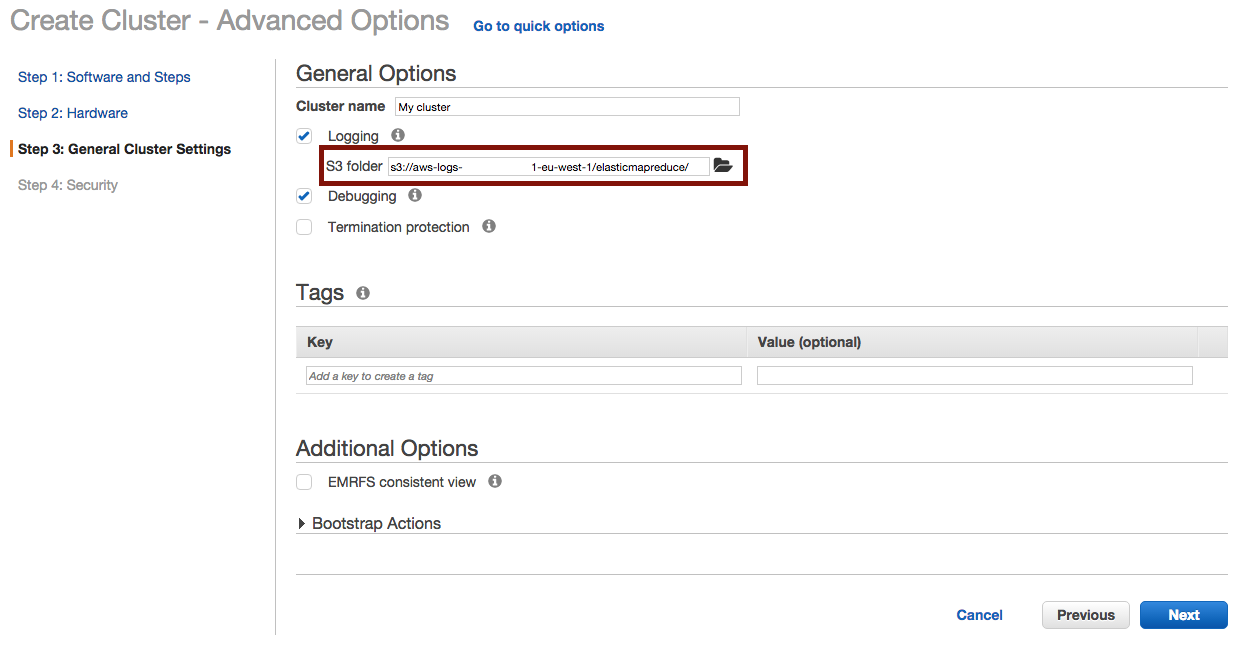
Once the cluster is terminated, you’ll find your logs in the choosen S3 folder at :

Giving the result in stdout.gz :
Opening s3://____.csv
Test Mean Squared Error = 11431.7565297
(we should have a better output in case of production, such as having the script upload the result to a specific well-named S3 folder instead) and the timings in stderr.gz :
RandomForest:
init: 24.770976383
total: 285.036716695
findSplitsBins: 1.594676031
findBestSplits: 259.45936596
chooseSplits: 259.178970344
As you can see, the computation of the random forest regressor lasted less than 5 minutes on 98 instances (instead of a few days on a single computer).
Well done!